The Potato Genome Sequencing Consortium (PGSC), an international team of scientists, focused on sequencing the genome of potato, has published its findings in the international journal Nature.
The Potato Genome Sequencing Consortium,initiated in January 2006 by the Plant Breeding Department of Wageningen UR (University &Research Centre) in the Netherlands, soon developed into a global consortium of 29 research groups from 14 countries.
Potato is the world's third most important food crop. It is a key member of the Solanaceae family of plants and a close relative of tomato, pepper, and eggplant. The potato genome sequence, the “genetic blueprint” of how a potato plant grows and reproduces, will assist potato scientists and breeders improve yield, quality, nutritional value and disease resistance of potato varieties, a process that has been slow in this genetically complex crop. The potato genome sequence will permit potato breeders to reduce the 10-12 years currently needed to breed new varieties. The potato genome is the first sequence of an Asterid to be published, group of flowering plants encompassing around 25% of all plant species.
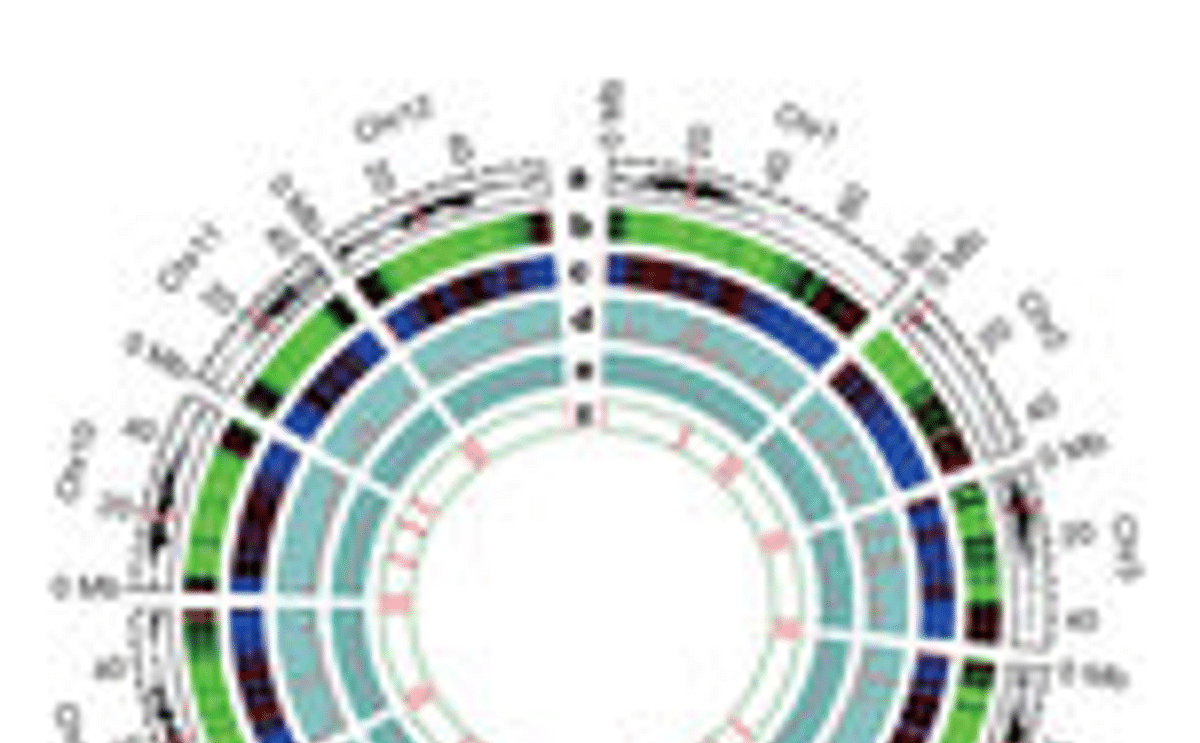
In late 2009, the Potato Genome Sequencing Consortium released a high quality draft sequence of the DM genome online. Since that time the PGSC has been refining the genome assembly, as well as performing exhaustive analysis and interpretation of the data. The genome assembly covers approximately 95% of the genes in potato, and was facilitated by new software developed by the BGI, one of the Chinese partners in the Potato Genome Sequencing Consortium.
Analysis of the genome sequence data has revealed that the potato genome contains approximately 39,000 protein coding genes. For over 90% of the genes the location on one of the 12 chromosomes is now known. The analysis also reveals that the potato genome has undergone extensive genome duplication though evolution.
Potato is an outbreeding crop plant, and comparisons of DM (double monoploid) and RH (diploid) data shed light on the phenomenon of inbreeding depression, from which potato suffers acutely. The data also show clear evidence for how expansion of particular gene families has contributed to the evolution of the potato tuber – the edible storage organ that is the most striking feature of this important and fascinating plant.
The potato genome assembly and other resources are now available in the public domain at www.potatogenome.net, where a complete listing and contact details for all Potato Genome Sequencing Consortium members can be found.
The Potato Council points out in a news release how this genome research can help the potato industry:
These recent developments in mapping the potato genome could offer growers access to varieties that are drought tolerant or are resistant to diseases such as blight or potato cyst nematodes.
“Genome mapping will facilitate innovative ways of normal breeding of potatoes as it will be easier to identify genes and genetic markers responsible for important variety traits” says Potato Council head of R&D, Dr Mike Storey.
“These findings will lead to a more in-depth understanding of potato biology and how the environment affects it. This could result in more efficient use of fertilisers, herbicides and pesticides, helping growers minimise cultivation costs whilst maximising sustainable yields.”
Dr Storey notes that although pests and diseases will continue to evolve, the new information on the genetic blueprint of the potato will help the sector develop responses to address these challenges.
The new knowledge is also expected to have significant impact on the amount of time it takes to develop a new variety. Characteristics are currently changed through conventional breeding programmes that take between ten and twelve years. It is expected that the genome sequence will provide a ‘street map’ for where the useful genes are, leading to faster development times.
“It is important for the potato industry to be ‘in tune’ with customers and the new technology will help it adapt more quickly to changes in consumer tastes,” continues Dr Storey.
“Potato Council, recognising these potential industry benefits from the project, contributed funding together with the Scottish Government, the Biotechnology and Biological Sciences Research Council and Defra. This enabled British researchers to make a significant contribution to the international assignment that comprised researchers from fourteen countries.”
Like to receive news like this by email? Join and Subscribe!
Join Our Telegram Channel for regular updates!
精选企业
Related News

十二月 22, 2024
Mooij Agro unveils innovative crop storage solutions at Potato Expo 2025

十二月 20, 2024
EU-Mercosur Free Trade Agreement: Reducing Tariffs and Impact on Agricultural Trade
Latest News
Sponsored Content
Sponsored Content
Sponsored Content
Sponsored Content
哪里
No location added
×
Sponsored Content







