Zebra chip causes discolouration in potato tubers, and processed potatoes.
CRC develops new DNA methods to detect zebrachip bacteria in potato psyllids

Accurate and timely identification of pest and disease threats is vital for the agricultural sector - the slower the response, the higher the economic cost, and the greater the likelihood a pest will spread.
Bacterial pathogens, including fireblight, citrus canker and annual ryegrass toxicity, present major biosecurity threats and are challenging to diagnose – current protocols are not able to identify bacteria at sub-specific levels of discrimination; e.g., pathovar, biovar, race, and may have an unacceptably high risk of false positives and false negatives.
In responding to this problem, Plant Biosecurity CRC scientists undertook the New Approaches for Diagnosing Bacterial Pathovars project. In one example of the work, the project team has identified stable regions in the genome of Candidatus Liberibacter solanacearum (CLso), which causes the serious disease Zebra Chip in potato and tomato crops.
CLso is particularly problematic to identify as it is non-culturable and vectored by psyllids. Its distribution within plant hosts is often erratic and contained, making detection by traditional diagnostic approaches of limited value. This is a huge problem for early detection.
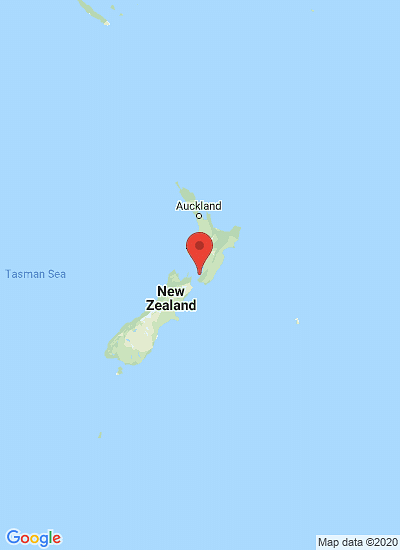
CLso was detected in New Zealand in 2008 and by 2012 this bacterium and the tomato potato psyllid, which transmits the pathogen, were estimated to have cost potato growers alone more than $200 million in crop damage. Recently the tomato potato psyllid was discovered for the first time in Australia in Perth Western Australia, including in horticultural areas to the north and south of Perth, causing damage to horticultural crops.
While the Department of Agriculture and Food WA indicated in June 2017 there "is no evidence of CLso at this time", the project scientists were able to provide significant science support to the WA incursion response team on the diagnostics on the bacterium in field samples.
During the project, Plant & Food Research scientists developed novel approaches to DNA extraction from individual psyllids, and using sophisticated bioinformatics programs assembled CLso genomes from samples that contained DNA from other organisms. The genome-informed and bioinformatics-enabled approach has a pathovar level of discrimination, and was validated for five bacterial pathogens including CLso by the project team.
This research will provide new diagnostics that will significantly reduce uncertainty in biosecurity, giving government and industry much faster and more accurate identification of plant pathogenic bacteria which are significant threats to agriculture, while reducing the false positives that interrupt trade and false negatives that decrease confidence in the system.
The research project New approaches for diagnosing bacterial pathovars is a collaboration between Plant Food Research (New Zealand), Department of Economic Development, Jobs, Transport and Resources Victoria, NSW Department of Primary Industries, Murdoch University and Kansas State University.
Bacterial pathogens, including fireblight, citrus canker and annual ryegrass toxicity, present major biosecurity threats and are challenging to diagnose – current protocols are not able to identify bacteria at sub-specific levels of discrimination; e.g., pathovar, biovar, race, and may have an unacceptably high risk of false positives and false negatives.
In responding to this problem, Plant Biosecurity CRC scientists undertook the New Approaches for Diagnosing Bacterial Pathovars project. In one example of the work, the project team has identified stable regions in the genome of Candidatus Liberibacter solanacearum (CLso), which causes the serious disease Zebra Chip in potato and tomato crops.
CLso is particularly problematic to identify as it is non-culturable and vectored by psyllids. Its distribution within plant hosts is often erratic and contained, making detection by traditional diagnostic approaches of limited value. This is a huge problem for early detection.
CLso was detected in New Zealand in 2008 and by 2012 this bacterium and the tomato potato psyllid, which transmits the pathogen, were estimated to have cost potato growers alone more than $200 million in crop damage. Recently the tomato potato psyllid was discovered for the first time in Australia in Perth Western Australia, including in horticultural areas to the north and south of Perth, causing damage to horticultural crops.
While the Department of Agriculture and Food WA indicated in June 2017 there "is no evidence of CLso at this time", the project scientists were able to provide significant science support to the WA incursion response team on the diagnostics on the bacterium in field samples.
During the project, Plant & Food Research scientists developed novel approaches to DNA extraction from individual psyllids, and using sophisticated bioinformatics programs assembled CLso genomes from samples that contained DNA from other organisms. The genome-informed and bioinformatics-enabled approach has a pathovar level of discrimination, and was validated for five bacterial pathogens including CLso by the project team.
This research will provide new diagnostics that will significantly reduce uncertainty in biosecurity, giving government and industry much faster and more accurate identification of plant pathogenic bacteria which are significant threats to agriculture, while reducing the false positives that interrupt trade and false negatives that decrease confidence in the system.
The research project New approaches for diagnosing bacterial pathovars is a collaboration between Plant Food Research (New Zealand), Department of Economic Development, Jobs, Transport and Resources Victoria, NSW Department of Primary Industries, Murdoch University and Kansas State University.
Like to receive news like this by email? Join and Subscribe!
NEW! Join Our BlueSky Channel for regular updates!
Sponsored Content
Sponsored Content
Sponsored Content
Sponsored Content
Sponsored Content