Scientific description of Brown Rot and Bacterial Wilt of Potato Caused by Ralstonia solanacearum (2020)
Based on Charkowski A., Sharma K., Parker M.L., Secor G.A., Elphinstone J. (2020) Bacterial Diseases of Potato. In: Campos H., Ortiz O. (eds) The Potato Crop. Springer, Cham
The authors of this content are Amy Charkowski, Kalpana Sharma, Monica L. Parker, Gary A. Secor, John Elphinstone
Brown Rot and Bacterial Wilt of Potato Caused by Ralstonia solanacearum
Taxonomy and Nomenclature
The genus Ralstonia is classified in the ß-Proteobacteria within the family Burkholderiaceae. The species complex of Ralstonia solanacearum has long been recognized as a group of phenotypically diverse strains, originally characterized as pathogenic races and biovars (Buddenhagen 1962; Hayward 1964).
More recently, Fegan and Prior (2005) described four phylotypes in the species complex, each comprising multiple phylogenetic variants (sequevars) according to sequence diversity within barcoding genes (including 16S rRNA, hrpB, mutS, and egl). Recently, the complex has been reclassified on the basis of whole-genome comparisons into three distinct species (Safni et al. 2014; Prior et al. 2016): R. solanacearum (Phylotype II), R. pseudosolanacearum (Phylotypes I and III), and R. syzygii (Phylotype IV) (image below).
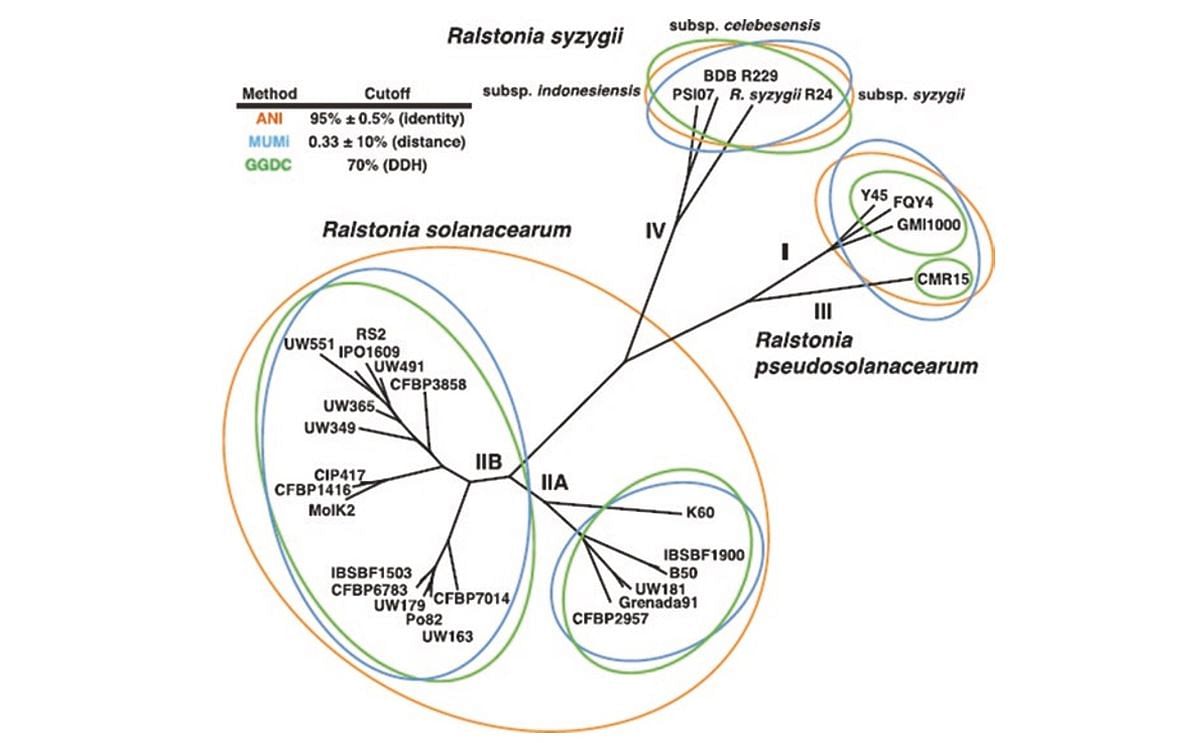
Phylogenetic relatedness of strains within the Ralstonia solanacearum species complex, from Prior et al. (2016). Concatenated tree of distance matrices generated from average nucleotide identity (ANI), maximum unique matches index (MUMi), and genome-to-genome distance calculator (GGDC) to compare DNA:DNA homology (DDH)
Host Range
In addition to potato (Solanum tuberosum and S. phureja), the large range of economically important hosts includes banana and plantain, cucurbits, eggplant, Eucalyptus, ginger, groundnut, mulberry, tobacco, tomato, and many ornamental plants. R. solanacearum, R. pseudosolanacearum, and R. syzygii each comprise strains that were originally designated as race 1 and which occur in tropical areas all over the world, attacking a very wide host range of over 250 hosts in 54 botanical families.
Some R. solanacearum genotypes within Phylotype II (sequevars IIA-6, IIA-24, IIA-41, IIA-53, IIB-3, IIB-4, and IIB-25), originally described as race 2, cause Moko disease of Musa spp. (banana and plantain) and Heliconia. R. syzygii comprises three subspecies: subsp. syzygii found on clove, subsp. celebesensis the cause of banana blood disease and subsp. indonesiensis found on solanaceaous crops (potato, tomato, and chilli pepper) as well as clove.
There are strains within each of the four phylotypes that can cause bacterial wilt and brown rot of potato; however, a single strain of R. solanacearum (sequevar 1) within Phylotype IIB (PIIB1), formerly known as race 3/biovar 2, is most widely associated with potato.
This genotype has a lower in planta temperature optimum (27 °C) than most other genotypes (35 °C), often occurring in latent (symptomless) infections at high altitudes in the tropics and in subtropical and temperate potato- growing areas. This strain can also cause bacterial wilt of tomato and can survive in perennial nightshades, which act as secondary hosts.
For example, the PIIB1 strain has overwintered in infected underground stolons of Solanum dulcamara (woody nightshade), growing along some European rivers, spreading to potato crops when the bacteria were transmitted in contaminated river water which was used for irrigation (Janse et al. 1998).
The same strain has also been spread internationally on geranium cuttings produced in Africa and Central America (Williamson et al. 2002).
Geographical Distribution
Phylotype I strains are regarded to be of Asian origin, Phylotype II strains are thought to be of South American origin, whereas Phylotype III appears to have evolved in Africa and Phylotype IV in Indonesia. The R. solanacearum species complex is widely designated as a quarantine organism in many countries in an effort to prevent its movement across geographical borders.
Nevertheless, the PIIB1 strain has spread from its origin to many potato-growing areas worldwide, presumably with movement in the trade of infected seed tubers (Elphinstone 2005). However, this strain has never been reported on potato in the USA, despite it having been introduced on infected geraniums (Williamson et al. 2002) and findings of other Phylotype II strains on potato and other hosts in the southern states.
The PIIB1 strain has in fact been designated as a select agent in the USA because of its perceived potential to pose a severe threat to agriculture.
Moko disease-causing Phylotype II strains mainly occur in South and Central America and the Caribbean, but also appear to have spread to the Philippines where the same strains have been found on cooking banana (ABB and BBB genotypes), causing the so-called bugtok disease.
Symptoms
Wilting is a common symptom of infections of most hosts with all phylotypes. The youngest leaves usually wilt first, appearing at the warmest time of day. Wilting may be visible in only one stem, on one side of a plant or even sectoral in part of a leaf, depending on where vascular infections are restricted in sectors of stems and leaf petioles.
Leaves may become bronzed or chlorotic and epinasty may occur. Wilting of the whole plant may follow rapidly if environmental conditions are favorable for pathogen growth. As the disease develops, a brown discoloration of the xylem vessels in the stem may be observed above the soil line and adventitious roots may develop.
A creamy, slimy mass of bacteria exudes from vascular bundles when the stem is cut. Wilting and collapse of whole plants can lead to rapid death. Symptoms on infected potato tubers may or may not be visible, depending on the state of development of the disease in relation to the prevailing temperature (image below).
Cutting a diseased tuber will reveal browning and necrosis of the vascular ring and in adjacent tissues. A creamy fluid exudate usually appears spontaneously from the vascular ring at the cut surface.
Bacterial ooze can emerge from the eyes and stem-end attachment of whole tubers, to which soil adheres. If cut stem or tuber vascular tissue is placed in water, threads of bacterial ooze exude.

Symptoms of potato brown rot with bacteria oozing from cut vascular tissues (a) and eyes (b) (UK Crown Copyright—Courtesy of Fera Science Ltd.), and wilted plant in the field (c) (Courtesy of International Potato Center)
Epidemiology
Although often described as soilborne pathogens, survival is usually short lived at low temperature in bare soil but is significant in alternative wild host plants (especially perennial nightshade species growing in waterlogged conditions or overwintering volunteers from susceptible crops).
The bacteria have been shown to survive in a viable but nonculturable (VBNC) form under stress conditions in soil and water (Kong et al. 2014), but the epidemiological relevance of this is unclear. Disease is usually most severe at temperatures of 24–35 °C, although the PIIB1 strain is more cold tolerant than other strains.
High soil moisture or periods of wet weather or rainy seasons are associated with high disease incidence. Entry into plants is usually through root injuries from where the bacteria move by colonization of the xylem where they adhere by polar attraction to the vessel walls, or invade the lumen.
Blocking of the vessels by bacterial extracellular polysaccharide (EPS) is considered to be the major cause of wilting. The bacteria can also be transmitted mechanically during pruning operations or when cuttings are taken for propagation. Long distance movement of vegetative propagating material (e.g. seed potatoes, rhizomes of ginger and turmeric, and banana suckers) can carry latent infections.
Natural infection of true seed has only been established for groundnut in Indonesia and China. There have been findings of contaminated seed of other hosts (including tomato, Capsicum, eggplant, and soyabean) although seed infection and transmission has not been substantiated.
At present, transmission through water or soil and movement of infected vegetative plant parts are considered to be more important for most host plants than transmission via true seed. In contrast, some strains of R. solanacearum and R. syzygii, which cause Moko disease and blood disease of banana and Sumatra disease of clove are transmitted by insects (including pollinating flies, bees, wasps, and thrips on banana and xylem-feeding spittlebugs of Hindola spp. on clove) with potential for rapid spread over several kilometers.
Pathogenicity Determinants and Resistance
Factors determining pathogenicity, virulence, and avirulence in the bacteria have been recently reviewed (Genin and Denny 2012; Meng 2013). After invasion into root intercellular spaces, expression of hrpB is induced, in response to plant signals, activating other hrp (hypersensitive response and pathogenicity) genes in construction (hrpK and hrpY) and regulation (hrpB and hrpG) of a type 3 secretion system (T3SS), a molecular syringe which is essential for pathogenicity.
The bacteria proliferate in intercellular spaces with the aid of a variety of effector proteins, secreted through the T3SS, which suppress plant defenses by interfering with host signal pathways.
HrpB positively regulates expression not only of hrp genes but also of genes encoding a number of plant cell wall-degrading exoproteins secreted through a type II secretion system (T2SS). These include polygalacturonases (PehA, PehB, and PehC), endoglucanase (egl), pectin methylesterase (Pme), and cellobiohydrolase (cbhA), which contribute not only to invasion of xylem vessels, leading to systemic infection, but also to quantitative control of virulence.
When cell densities reach a threshold, PhcA is activated by build-up of a volatile quorum sensing signal, 3-hydroxy palmitic acid methyl ester (3-OH PAME), inducing genes (epsABCDEF & P) controlling biosynthesis of exopolysaccharide (EPS), a major virulence factor.
Transcriptome analysis (Jacobs et al. 2012) has also confirmed expression of these and other genes encoding various virulence traits at high cell densities during host infection, including genes imparting stress tolerance (bcp, acrA, acrB, and dps) and motility and attachment structures (pilA and fliC).
Available field resistance to the R. solanacearum species complex is limited and tends to be unstable under different environmental conditions and/or strain variability.
Traditional breeding has not yet yielded new resistant varieties because of the difficulty in transferring multiple unknown genes from wild germplasm with polygenic resistance into cultivars without cotransferring undesirable linked traits.
Furthermore, high-level resistance to host colonization as well as to disease development is needed to avoid the risk of spreading the pathogens in symptomless latent infections. Most studies on the genetic basis of resistance to bacterial wilt have been conducted in the model plants Arabidopsis thaliana and Medicago truncatula (Huet 2014).
Quantitative trait loci (QTL) have been identified that include R genes that encode proteins that recognize bacterial effector avirulence (AVR) proteins, triggering resistance to the bacterium. Transfer of selected R genes from A. thaliana into tomato conferred immunity to the Ralstonia strain with the corresponding AVR gene (Narusaka et al. 2013).
Both broad-spectrum and strain-specific quantitative trait loci (QTLs) have been identified in tomato (Wang et al. 2013), tobacco (Qian et al. 2013) and eggplant (Salgon et al. 2017).
Discovery of possible resistance/ avirulence (R/Avr) gene for gene resistance mechanisms is particularly interesting since known bacterial effectors can be used to screen for homologous resistance genes in related crops, including potato.
Management
Disease management remains limited and is hampered by the ability of the pathogens to survive in wet environments on plant debris or in asymptomatic weed hosts, which act as inoculum reservoirs. In the absence of any curative chemical control methods, prevention of bacterial wilt largely relies on the availability of pathogen- free planting material and effective surveillance and monitoring to protect areas free from the bacteria.
For potato, effective disease management has mainly resulted from the use of limited generation seed multiplication from pathogen-free nuclear stocks with zero tolerances for the disease in official seed certification programs.
Regular post-harvest testing of seed potato tubers is usually also necessary to avoid distribution of latent infections. Similarly, for other vegetatively propagated crops, there is a need to ensure planting material has been tested free of infection and that there are restrictions on the movement of planting material from affected to disease- free areas.
Disinfection of pruning and harvesting tools is important in preventing spread of disease e.g. in banana and plantain production. In areas where the pathogen could be spread in contaminated irrigation water, prohibition of irrigation with surface water has been an effective control measure.
For hydroponic glasshouse production systems, the disinfection of recirculating water (e.g. using chlorine dioxide) can prevent spread of the bacterium. This effectively halted international spread of R. solanacearum PIIB1 in geranium cuttings produced in Central America and East Africa following export to the USA and Europe.
